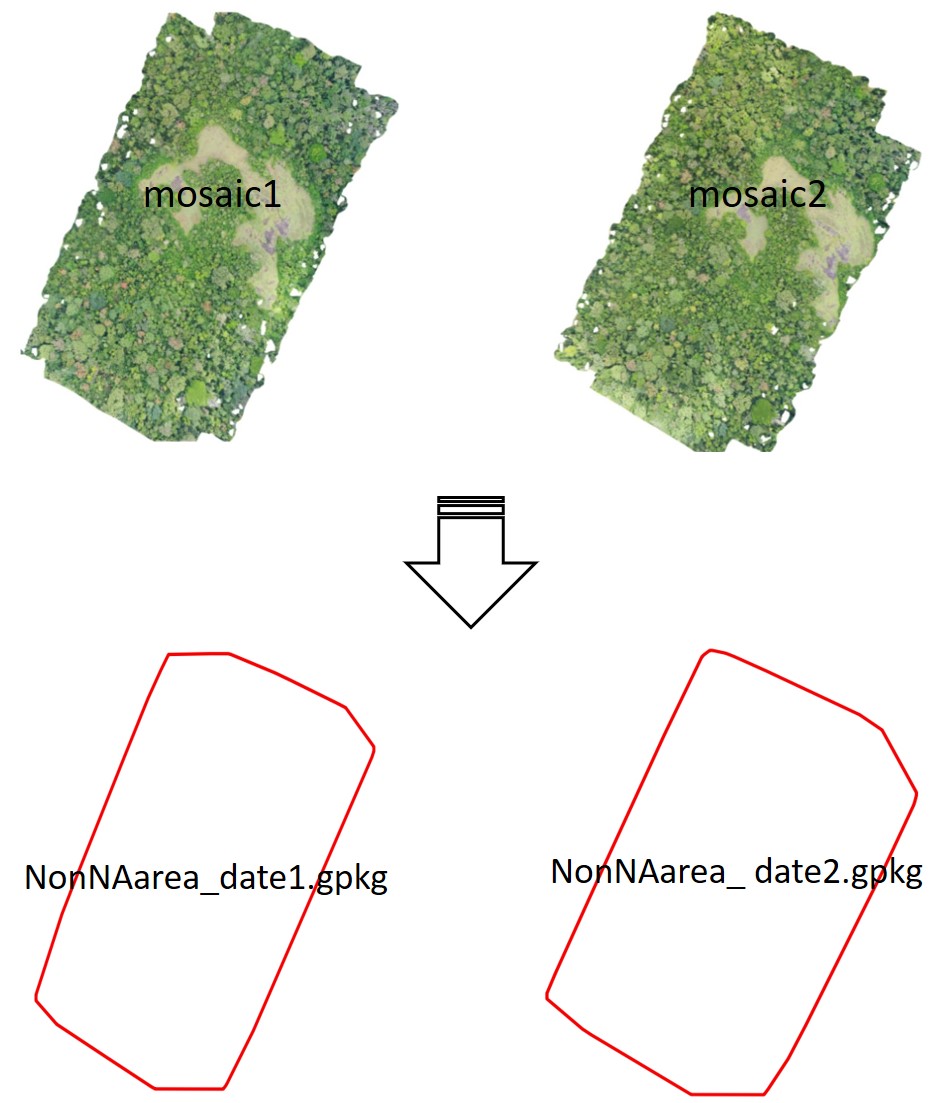
check_crownsFile(path_crowns = file.path(my_path, "3_crowns/crowns_2024_11_04.gpkg"))
sf::read_sf(path_crowns) %>%
rename(
geometry = geom,
family = tax_fam,
genus = tax_gen,
species = tx_sp_lvl,
code_sp = idtax_f
) %>%
sf::write_sf(.,file.path(my_path, "3_crowns/crowns_2024_11_04_rename.gpkg"))
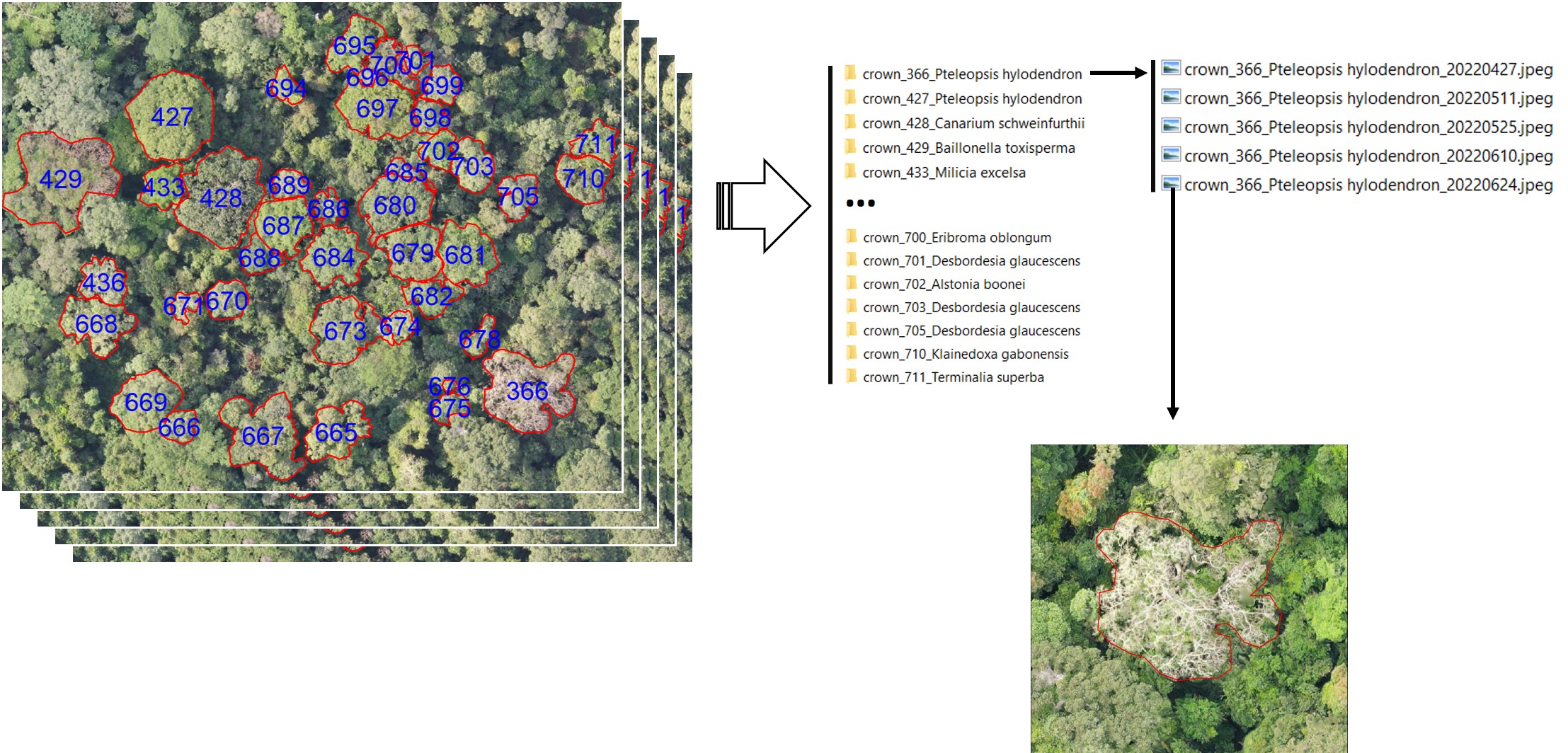
extract_crownsImages(
path_in = file.path(my_path, "5_arosics_output"),
path_crowns = file.path(my_path, "3_crowns/crowns_2024_11_04_rename.gpkg"),
path_bbox = file.path(my_path, "6_bbox"),
out_dir_path = file.path(my_path, "7_crownsImages"),
site = NULL,
dates = NULL,
N_cores = 1,
width = 720,
height = 825
)
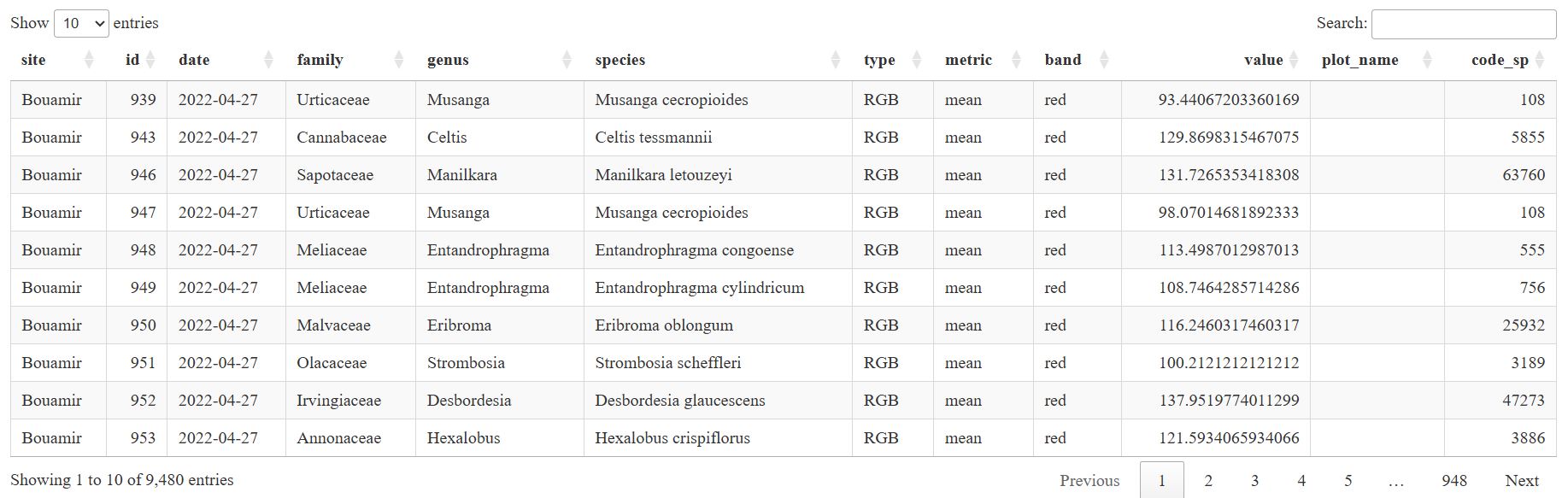
rgb_data <- extract_rgbValues (
path_in = file.path(directory, "5_arosics_output"),
path_crowns = file.path(directory, "3_crowns/crowns_2024_11_04_rename.gpkg"),
out_dir_path = file.path(directory, "9_outputs/"),
ncor = 1,
sites = NULL,
dates = NULL
)
pivotLabels <- pivot_Labels(labels_path = file.path(directory, "8_labeling_file/Bouamir_labelingFile.xlsx"),
out_dir_path = file.path(directory, "9_outputs"),
simplify_labels = TRUE)
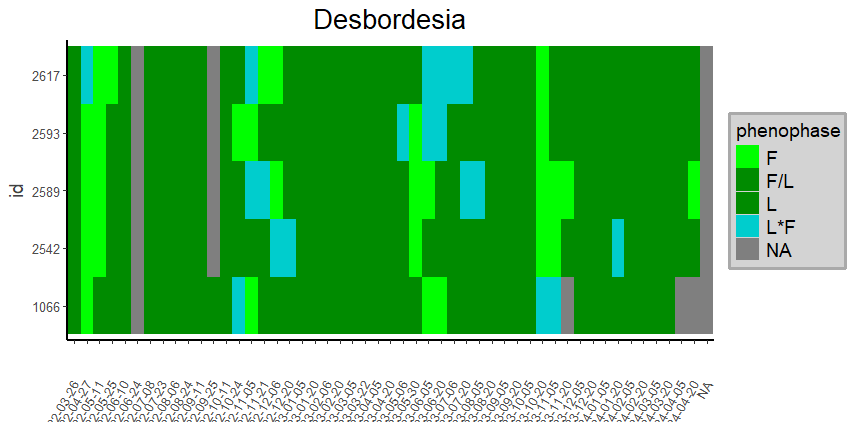
heatmap_Labels(pivotLabels,
Specie = NULL,
Genus = 'Desbordesia',
Family = NULL,
title = NULL)
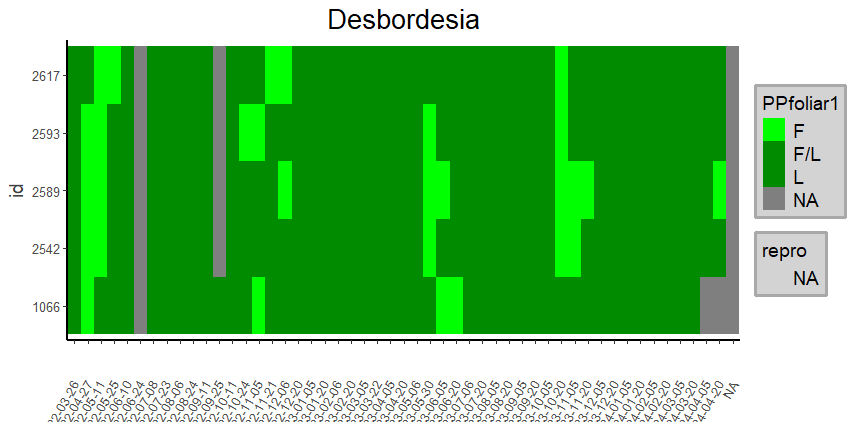
heatmap_Labels(pivotLabels,
Specie = NULL,
Genus = 'Desbordesia',
Family = NULL,
title = NULL,
simplify = TRUE)
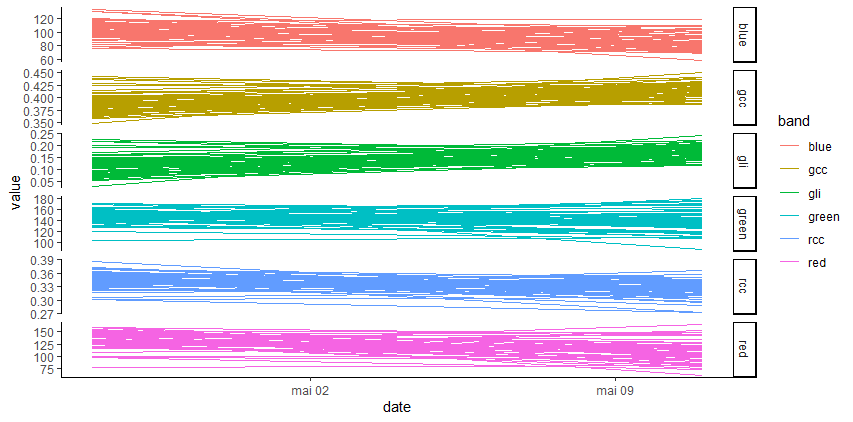
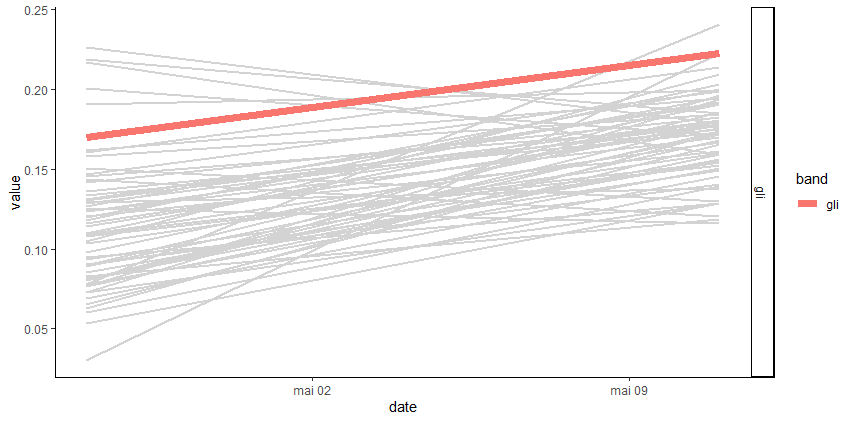
plot_signal(data = merge_data, Genus = 'Desbordesia', slcted_id = 2614)
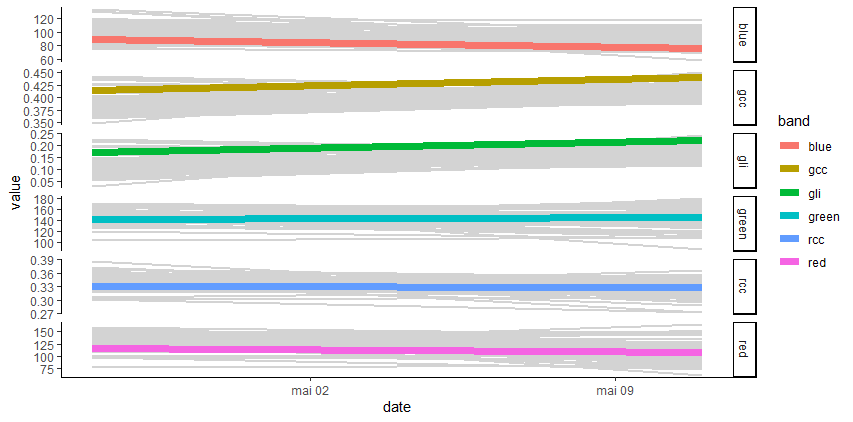
plot_signal(data = merge_data, Genus = 'Desbordesia', slcted_id = 2614, Band = c('gli', 'gndvi'))